Label-Free Nanoscale-Sensitive Study of Sub-Cellular Morphology
Despite the fact that cancer is a disease caused by genetic abnormalities, the most widely accepted means of diagnosing and characterizing cancer cells is with H&E stained microscopic imaging. The identifying features
of cancer include abnormally large and crowded cell nuclei with altered cellular organization and shape and increased number of nucleoli. Alterations in other organelles including mitochondria, lysosomes, Golgi bodies,
and vesicles are also observed in cancer when the corresponding stain is used to mark each organelle. In addition, changes in nuclear substructures are studied as many of them are directly involved in genetic transcription.
Though genetic alterations can also be identified in cancer cells, the implications of these genetic alterations are often more difficult to interpret than a direct image of the cellular phenotype.
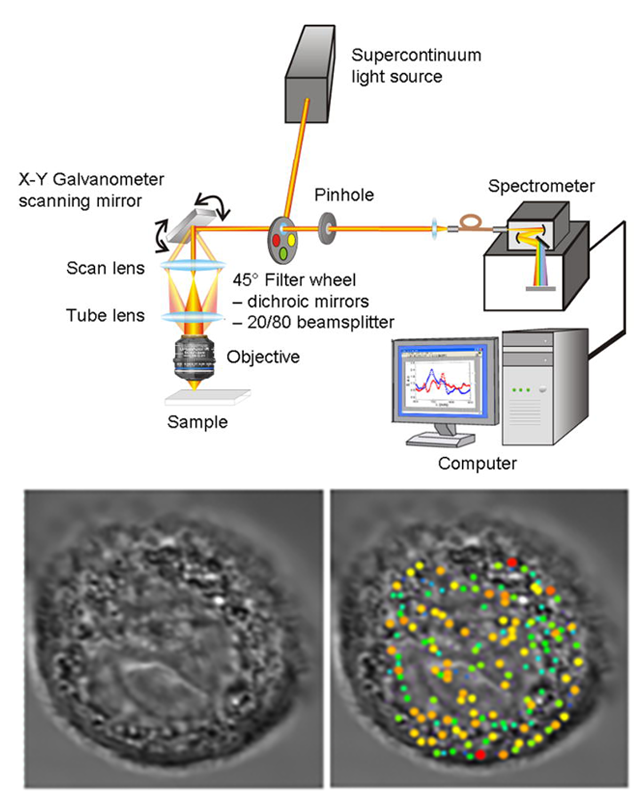
While cellular alterations in organelle and nuclear structure are readily observed and studied in cancer, there are fundamental limitations in existing imaging techniques that prevent the study of very early stage pre-cancerous
alterations. Perhaps the most often used imaging tool for observing cellular structure is fluorescence laser scanning confocal microscopy. Although fluorescence microscopy can achieve targeted contrast for specific
organelles or proteins, imaging in living cells remains limited to just a few types of fluorophores and therefore structure types. Perhaps an even more substantial limitation of fluorescence microscopy and most other
conventional optical imaging is that they are subject to the diffraction limit and cannot discern the properties of cellular structures that are significantly smaller than the wavelength. On the other hand, high resolution
electron microscopy imaging methods are destructive, involve extensive manipulations with the sample, and cannot be utilized in living systems.
In order to overcome the limitations of both optical and electron
microscopy, an imaging method that is based on an entirely different physical principle is required. This method should ideally identify all important cellular structures in live cells, while simultaneously characterizing
them with sub-diffraction accuracy. In 2007 we introduced confocal light absorption and scattering spectroscopic (CLASS) microscopy, a new native contrast imaging technique based on physical principles of light scattering,
which is capable of noninvasively determining the dimensions and other physical properties of single subcellular organelles and other structures in live cells at sub-diffraction spatial scales and quantifies their physical
properties when cells undergo pre-cancerous alterations, by using light scattering spectra as native optical biomarkers. The CLASS technique allows for the reconstruction of the scatterer size map, which can be overlaid
on the bright-field cell image, providing information about the positions of various organelles and other structures inside the cell. CLASS provides unique capabilities to study functions of viable cells, which are
beyond the capabilities of other techniques (PNAS 2007).